The low density lipoprotein receptor (LDLR) gene family consists of cell surface proteins involved in receptor-mediated endocytosis of specific ligands. Low density lipoprotein (LDL) is normally bound at the cell membrane and taken into the cell ending up in lysosomes where the protein is degraded and the cholesterol is made available for repression of microsomal enzyme 3-hydroxy-3-methylglutaryl coenzyme A (HMG CoA) reductase, the rate-limiting step in cholesterol synthesis. At the same time, a reciprocal stimulation of cholesterol ester synthesis takes place. Mutations in this gene cause the autosomal dominant disorder, familial hypercholesterolemia. Alternate splicing results in multiple transcript variants.[provided by RefSeq, Sep 2010]
- Lipid metabolic process
- Lipid transport
- Endocytosis
- Receptor-mediated endocytosis
- Phagocytosis
- Hypercholesterolemia, familial, 1
- Homozygous familial hypercholesterolemia
- Familial hypercholesterolemia
- Carotid artery dissection
- Carotid artery occlusion
- Familial Hypercholesterolemia
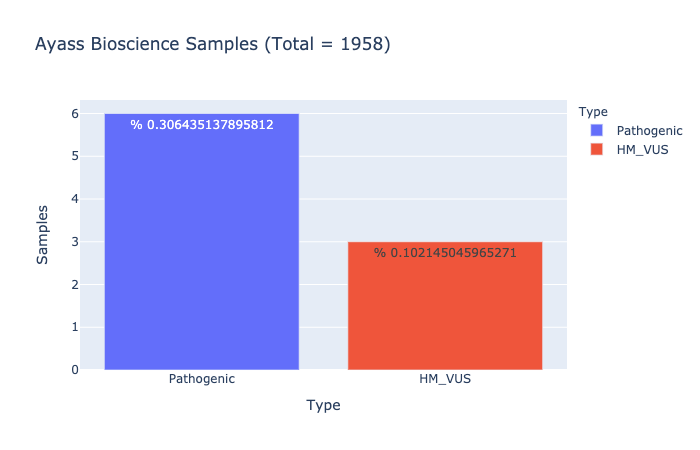
Based on Ayass Bioscience, LLC Data Analysis
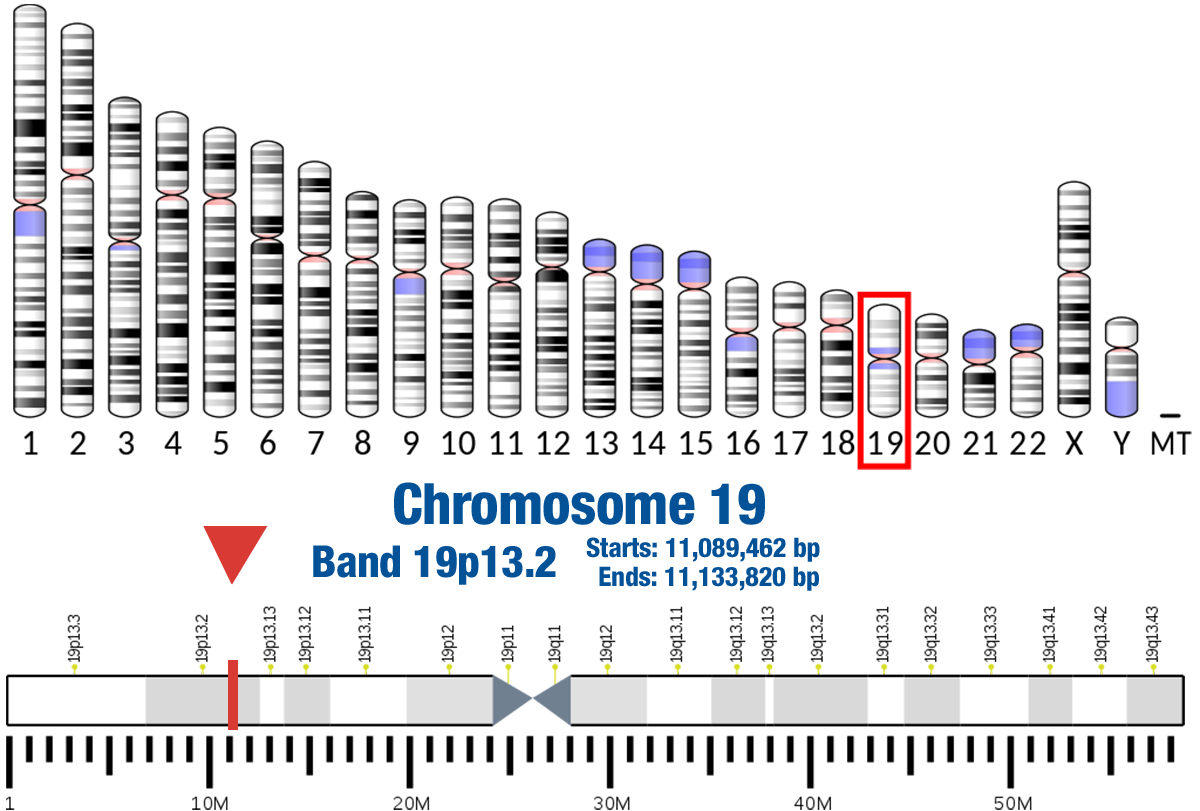
Gene Location
Pathogenic Prevalence
% 0.306435137895812
Ratio of samples with at least 1 pathogenic variant (Computed from Ayass Bioscience Samples)
HM-VUS Prevalence
% 0.102145045965271
Ratio of samples with at least 1 High/Med VUS variant (Computed from Ayass Bioscience Samples)
Pathogenic Variants
C=0.000024(3/125568,TOPMED)
T=0.00003(1/31390,GnomAD)
A=0.0000(0/2188,ALFAProject)
C=0.0000(0/2188,dbGaPPopulationFrequencyProject)
T=0.0000(0/2188,dbGaPPopulationFrequencyProject)
A=0.000028(7/251066,GnomAD_exome)
A=0.000008(1/125568,TOPMED)
A=0.000025(3/120814,ExAC)
A=0.00003(1/31388,GnomAD)
A=0.0000(0/3854,ALSPAC)
A=0.0003(1/3708,TWINSUK)
A=0.0000(0/2188,ALFAProject)
T=0.000056(14/251206,GnomAD_exome)
T=0.000082(10/121250,ExAC)
T=0.00003(1/31372,GnomAD)
C=0.00000(0/78688,PAGE_STUDY)
T=0.0001(1/8988,ALFAProject)
T=0.0000(0/3854,ALSPAC)
T=0.0003(1/3708,TWINSUK)
High/Med VUS Variants
T=0.000012(3/250932,GnomAD_exome)
T=0.000017(2/119950,ExAC)
T=0.0001(1/8988,ALFAProject)
C=0.000163(41/251310,GnomAD_exome)
C=0.000151(19/125568,TOPMED)
C=0.000100(12/120588,ExAC)
C=0.00006(2/31400,GnomAD)
C=0.0000(0/2188,ALFAProject)